Induced pluripotent stem cells (iPSCs)
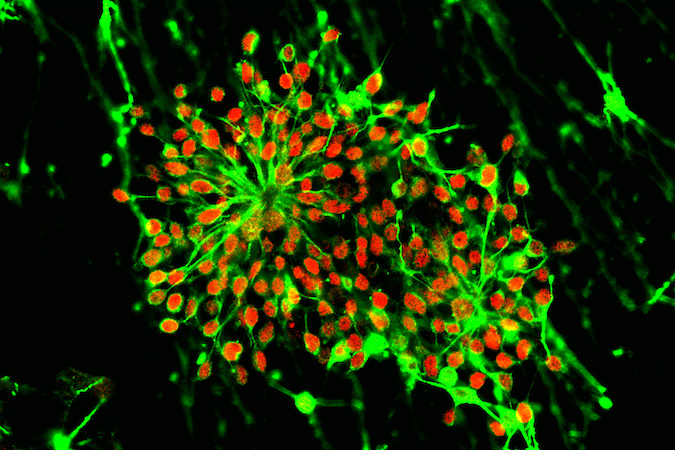
iPSCs are generated using the latest techniques to reprogram, expand and characterize human iPS cells from human skin or blood tissues of healthy subjects and diseased patients. We then turn the iPS cells into specific cells of the human body, including components of the nervous system for use by researchers. Motor neurons (iMNs) are generated from human induced pluripotent stem cells (iPSCs) from patients with ALS and SMA and for unaffected controls. The iPSC core facility at Cedars-Sinai Medical Center (CSMC) has coordinated iPS cell expansion and differentiation protocols to ensure consistency across sites. We have developed standard operating procedures (SOPs), QC measures, and an improved protocol for the maintenance and differentiation of the cells.
Perturbations
The genetic disease background in iPSC lines from individuals with SMA (SMN1 mutation), C9ORF72 repeat expansion associated ALS (C9-ALS) and sporadic ALS are genetic perturbagens associated with relevant iPSC-derived CNS cell types we will generate (e.g. Motor neurons). Importantly, additional chemical perturbagens will be selected based on their applicability to a wide range of cell types and cellular signaling pathways.
Proteomics
We perform SWATH-MS (Sequential Window Acquisition of all THeoretical fragment ion spectra by Mass-Spectrometry), a data-independent acquisition (DIA) method. DIA library generation, required for the primary method of analyzing DIA data, can be performed by several methods and there is no current standard in the field. We tested several methods and variables that go into generating an iPSC and iMN specific library, including data generated on multiple different instruments, various alterations within the instrument method itself and different programs that use the data to make the library. Sample specific libraries have been generated from aliquots of all iPSC and iMN samples respectively to generate an iPSC and iMN library.
Epigenomics
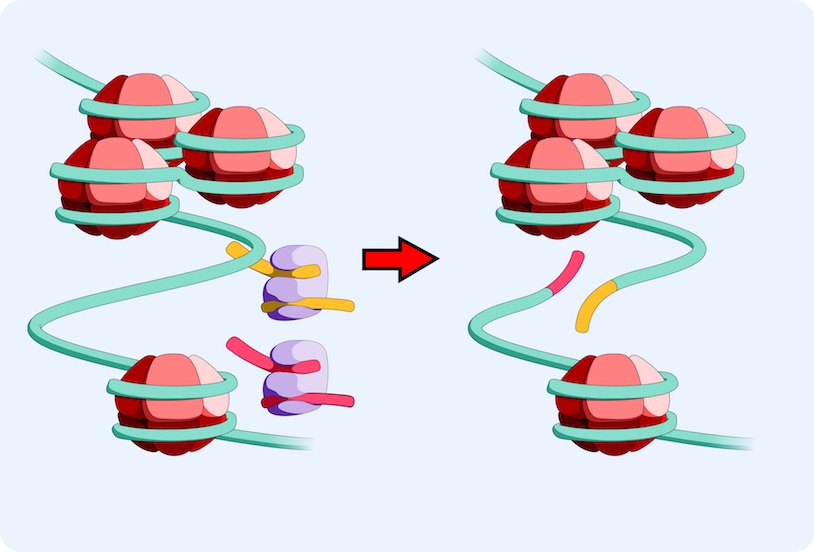
ATAC-Seq is a relatively new transposase-based, deep sequencing based epigenomic assay used to assess chromatin accessibility and identify functional regulatory sites involved in driving transcriptional changes associated with cell responses to perturbations. ATAC-Seq detects open chromatin sites and maps transcription factor binding events in regulatory elements genome-wide, without needing any prior information about which proteins are bound. ATAC-Seq signals allow for the delineation of fine-scale architectures of the regulatory framework by correlating occupancy patterns with other features, such as global gene induction programs.
Robotic Imaging
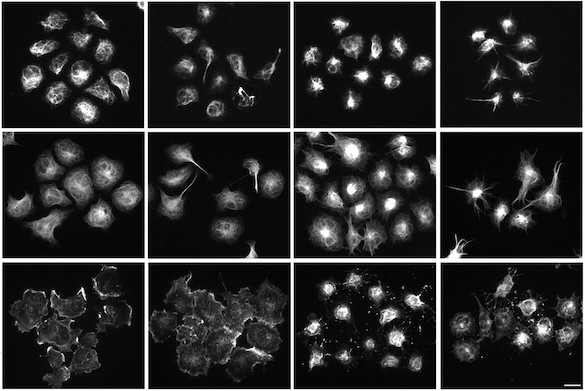
Automated robotic microscopy (RM) is used to identify and track live individual neurons in a high throughput and high content fashion over time. Automated image analysis is used to quantify intermediate changes in the physiology of a given cell and relate it to that cell’s fate. From these measurements, mulivariate predictive dynamic models of cell fate are constructed that weigh co-variates based on the magnitude and nature of their predictive power. These models offer a signature of the cell’s biology and a blueprint for rational therapeutic interventions.